Advised by HIlary Coller
May 7, 2008
As my second semester of independent work for the Computer Science Department, I developed a more effective method of determining levels of a certain cellular process known as autophagy from images.
My advisor, Hilary Coller, was interested in finding a more reliable means of quantifying the amount and type of cellular autophagy occuring in different cells. Several objectives were established for this program. The first goal was to develop something that worked reliably. That is to say that the program must quantify equally those cells experiencing the same levels of autophagy regardless of other conditions relating to the cells, or the image itself. While this may seem trivial it was a significant computational problem.
The second goal of the project was to develop a program that would be transferrable to other areas of biology. There are a large number of problems in image analysis that require the quantification of some individual segment of total cellular information. As a result, it would be nice if this program could, with minor modifications, be made to solve those problems as well.
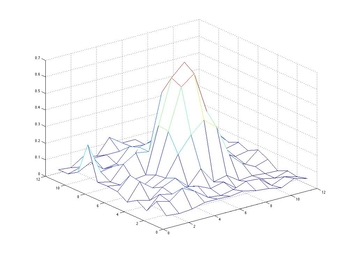
The results of this project are shown in the paper at right. In general, these findings were positive. Even with some simple heuristics, I was able to quantify and categorize cellular autophagy. In addition, I successfully attempted to run this program on a number of other data sets taken either from different resolution microscopes or even using different samples or organisms.
I would like to thank Hilary Coller for her help on this project.